研究概要
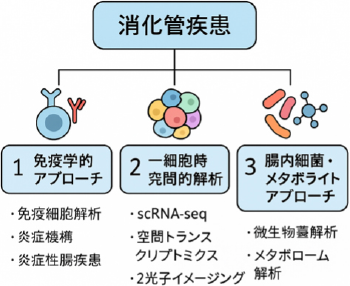
消化器疾患は、管腔臓器である食道・胃・小腸・大腸、および実質臓器である肝臓・膵臓など多岐にわたる臓器に発症し、がん、炎症性疾患、機能性疾患といった多様な病態を呈する。
これまでに多くの治療法が開発されてきたものの、疾患の定義が主に症状や内視鏡的形態変化などのマクロな所見に依存しており、病態の本質的理解には至っていない。加えて、同一診断名の患者間で薬剤反応性が大きく異なるなど、疾患の多様性(heterogeneity)が臨床的課題となっている。
本研究室では、消化器疾患の発症・進展メカニズムを分子・細胞レベルで明らかにすることを目的に、以下の3つのアプローチを統合的に用いて解析を行う。
- 1. 免疫学的解析による病態解明
- 消化管は、腸管免疫系を中心に極めて多様な免疫細胞群が集積する免疫臓器でもあり、免疫細胞間のネットワーク破綻は多くの炎症性疾患・腫瘍性疾患の発症に関与すると考えられている。患者由来検体を用いて、免疫細胞の構成、活性化状態、相互作用の解析を行い、免疫の失調が病態に与える影響を明らかにする。
- 2. 一細胞・空間トランスクリプトーム解析およびライブイメージング
- 近年発展した single cell RNA-seq 、空間トランスクリプトミクス(spatial transcriptomics)、2光子励起顕微鏡(2-photon imaging)等の技術を活用し、組織内の細胞分布とその転写プロファイルを高次元かつ空間的に解析する。さらに、動態観察により免疫細胞や上皮細胞の局在・移動・相互作用をリアルタイムに観察し、組織階層性における細胞機能の時空間的理解を図る。
- 3. 腸内細菌叢およびメタボローム解析による代謝・免疫連関の解明
- 腸内細菌叢(マイクロバイオータ)は、宿主免疫の成熟、粘膜恒常性維持、代謝制御に深く関与しており、近年その代謝産物(短鎖脂肪酸、バイオアミン類など)が疾患発症に関与することが示唆されている。本研究では、患者由来の糞便検体を用いた16S rRNAシークエンスやメタゲノム解析、ならびにメタボローム解析により、腸内細菌と宿主の病態形成の相互作用を解明する。
- 研究の展望
- これら3つのアプローチを統合的に活用することで、消化器疾患の病態を多階層的に解析し、将来的には個別化医療や新規治療標的の創出に貢献することを目指す。
Research Overview
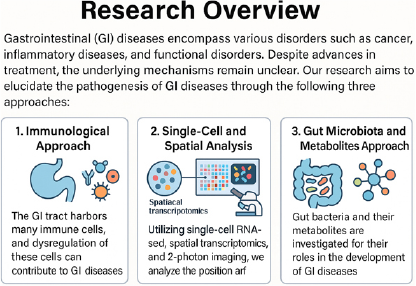
Gastrointestinal (GI) diseases encompass a wide range of conditions affecting both hollow organs (esophagus, stomach, small intestine, colon) and solid organs (liver, pancreas). These include cancer, inflammatory diseases, and functional disorders. Although numerous therapeutic strategies have been developed, disease definitions are still largely based on macroscopic findings such as symptoms and endoscopic morphology. This has limited our ability to stratify patients biologically and has led to variable treatment responses among individuals. To elucidate the underlying mechanisms of these diseases at molecular and cellular levels, our lab propose a multifaceted approach incorporating the following three methodologies:
- 1. Immunological Analysis of GI Diseases
- The gastrointestinal tract is rich in immune cells, especially within gut-associated lymphoid tissue (GALT). Dysregulation of immune cell interactions is thought to be central to the pathogenesis of many inflammatory and neoplastic GI diseases. We analyze patient-derived samples to investigate immune cell composition, activation status, and interactions, aiming to clarify the role of immune dysregulation in disease.
- 2. Single-cell and Spatial Transcriptomic Analysis with Live Imaging
- Advanced technologies such as single-cell RNA sequencing (scRNA-seq), spatial transcriptomics, and two-photon intravital imaging enable us to investigate the transcriptomic profiles and spatial organization of individual cells within tissue. In combination with live imaging, this allows real-time tracking of immune and epithelial cell behavior, offering insight into the spatiotemporal orchestration of cellular functions during disease progression.
- 3. Microbiome and Metabolite-based Approaches
- The intestinal microbiota plays a critical role in immune regulation, mucosal homeostasis, and host metabolism. Recent studies have revealed that microbial metabolites (e.g., short-chain fatty acids, biogenic amines) are key modulators of disease states. We perform 16S rRNA sequencing, metagenomics, and metabolomic profiling of patient-derived stool samples to elucidate the host–microbiota–metabolite axis in GI pathology.
- By integrating these three approaches, we aim to decode the complex pathophysiology of gastrointestinal diseases and contribute to the development of precision medicine and novel therapeutic targets.

